ML: My First with Parallel Computing
Parallel computing is fun, but quiet different!
Intro
“Big data is like teenage sex: everyone talks about it, nobody really knows how to do it, everyone thinks everyone else is doing it, so everyone claims they are doing it…” — Dan Ariely
And the same quote can apply to many other related terms with big data like machine learning, deep learning, data visualization and parallel computing. Fortunately, I did get a chance to do one of them recently.
How I Get Into This Whole Parallel Computing Business?
Over the pass summer, I have been doing some research and implemented an approximate algorithm for efficient graph matching. Utilizing this algorithm, I later developed a spatial pattern matching model following the research of my mentor Prof. Hong.
Even though the graph matching runs relatively fast, it still takes about 30 mins to match two graphs with about 100 nodes and such matching is needed to run several rounds in our model developing algorithm. As bad as it sounds, we were actually quiet happy with our progress at that point and we were able to demonstrate the viability of our model through small sample set.
I mean, every computer science problem is not a problem until we need to scale.
But,
Going into this fall, we decided to apply the algorithm to biology domain and aim to solve a fascinating yet challenging problem: generate a model to learn the common domain across all the sample proteins. We convert sample protein crystal structures into graphs and hope to understand the common structure in this sample protein utilizing our algorithm. This is a fascinating job. I mean, look at this beautiful matching graph:
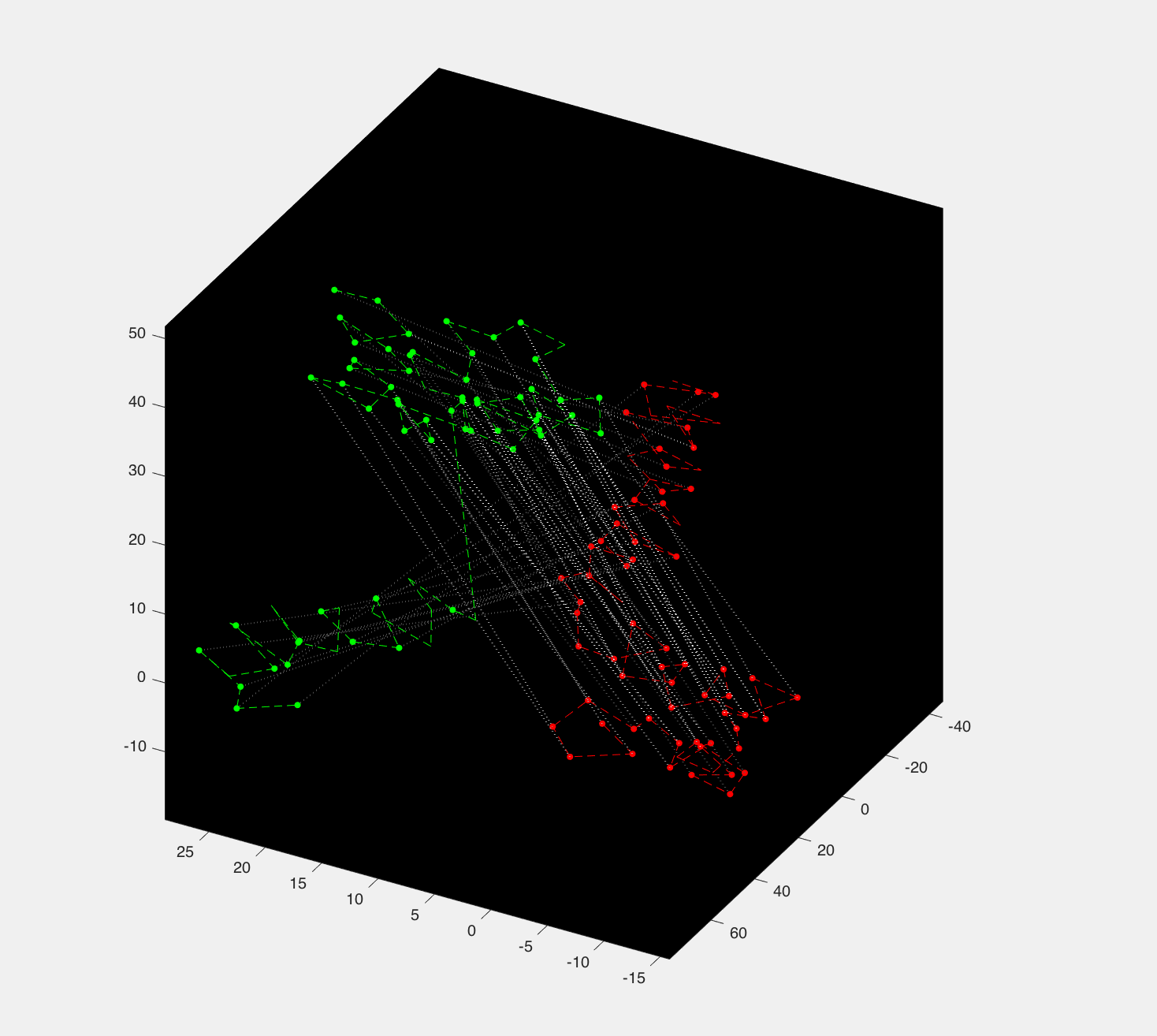
But we run into a problem: the scaling problem. Most of the proteins have hundreds of amino acids and give us a graph with hundreds of nodes. Therefore, we need to make our graph matching faster. Since the majority time for running the graph matching algorithm is used to build a huge matrix storing the edge compatibility between all the edges of two graphs. The work can be huge once we scale. Let’s say we have a graph with A nodes and another graph with I nodes. Then there will be at most A*A edges in the first graph and I*I edges in the later. Therefore we will need to compute a huge matrix with size (A*A)*(I*I). That’s a lot of work, but luckily the computing order does not matter, so we decide to try parallel computing.
The Parallel Computing Toolbox in MATLAB
MATLAB has this amazing parallel computing toolbox that can help you utilizing multiple CPU on your local machine or clusters to do parallel computing by simply change your for loop to parfor loop. That sounds pretty straight forward and that’s what I thought when I first looked at the tutorial.
However, the complication in parfor is the way we can index our matrix. It is limited. Here are some legal cases if we want to assign a value to our (A*A)*(I*I) matrix C.
C(2,7)=1;
C(p,5)=1;
C(p,:)=1;
And here are some illegal cases:
C(p,a)=1; % a is some random variable
C(p,p)=1;
C(p,1:a)=1;
C(p,1:p)=1;
So as you can see, to compute a matrix using parallel computation, MATLAB requires you to compute it by a whole row/column or just a constant cell.
This sounds easy, but in many cases, it requires us to transform our representation or even conceptualization of the problem.
In my case, I partition my (A*A)*(I*I) matrix C into A*I blocks and compute these blocks one by one using two nested for loop. However, I cannot do this in the new parfoor loop since we need to fill out the matrix with a whole row. I had a really hard time conceptualizing the process in the beginning because I think of my matrix blocks instead of rows or columns. Now that we need to partition the matrix by row, everything looks so different now and I am not sure how to conceptualize such process.
I eventually solve this problem by drawing out the whole matrix and it is actually quiet simple. But I learn a lot from this process.
Why parallel computing is fun? (And what I learn?)
Since parallel computing is doing the computation in parallel manner, it requires a different mindset compared to sequential computing.
The first thing is how we organized our data structure. If we are using parallel toolbox/library, many of them require us to slice our variable in a column/row manner to fit their implementation, just like in MATLAB. To deal with this, we might need to tweak the data structure in our algorithm and some times that can be counter intuitive.
Let’s look at a matrix in Dijkstra Algorithm:

As we can see, we are growing the matrix from bottom left to top right and parallel computing can actually be utilize to compute each element in diagonal layer. In that case, we will need to map our original matrix into a different matrix so that we can slice diagonal layer into column/row. Such matrix can be in weird shape and hard to conceptualize.
Moreover, we need to be extra careful about racing condition. In my case, the computation is just static mapping and values in C does not involve in the computing process, so it is safe to run the computation in whatever order the system wants. But in many other cases, this is not true and we might need extra instructions to implement the synchronization.
This sounds complicated, but with all the toolboxes out there, we are able to build parallel computing solution in a couple of hours and the improvement is amazing:
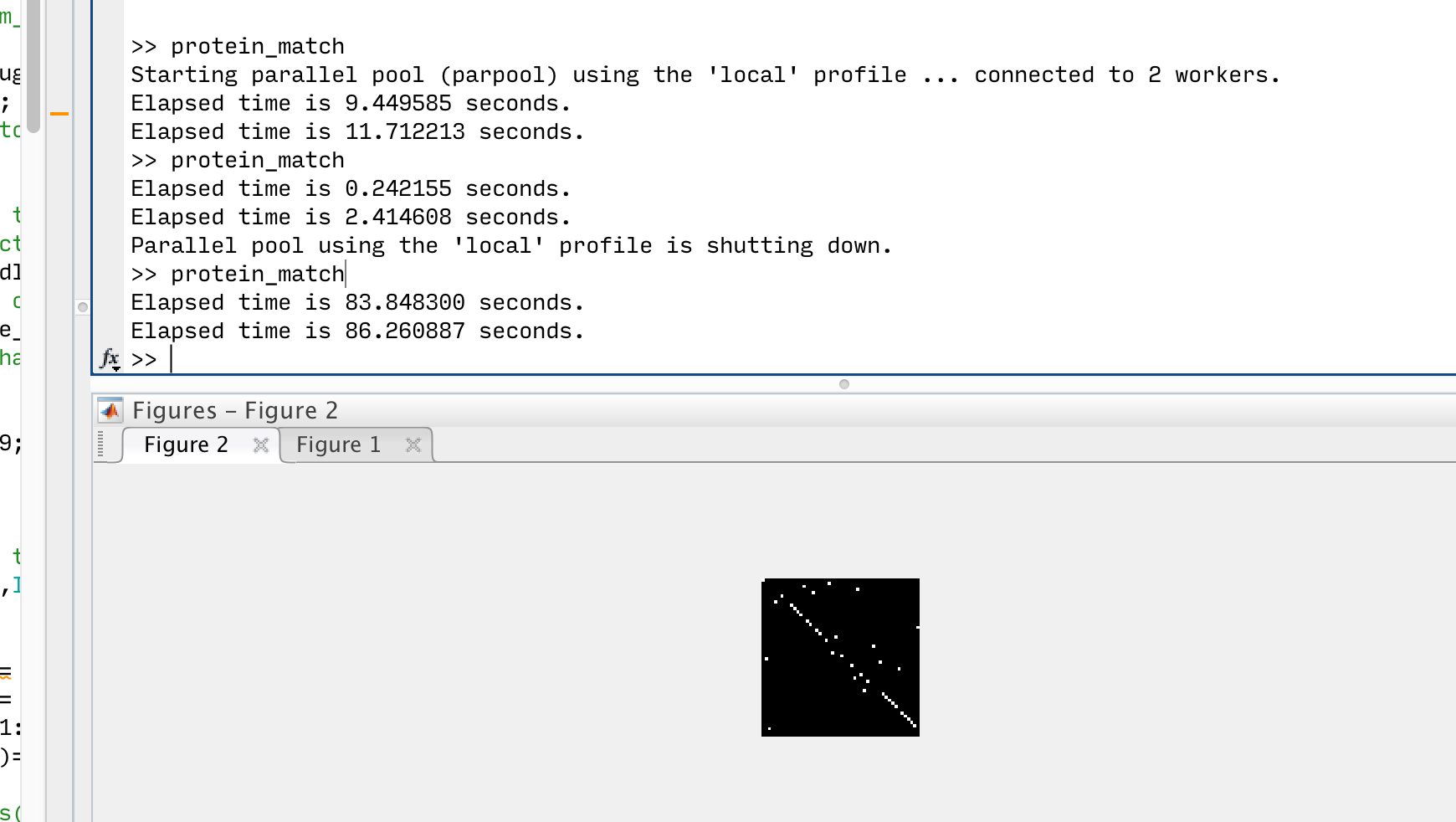
For me, MATLAB first start pooling “workers” for parallel computing works and this takes some time. Once the pooling is done in the first round, it only takes 0.2 seconds to finish a round of edges compatibility computing, while the same process takes 84 seconds without parallel computing. So I literally jumped out of my bed when it actually works!
And yep, I code on my bed too. :)
